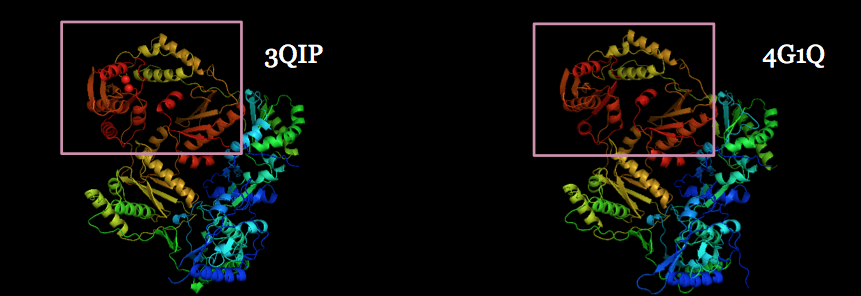
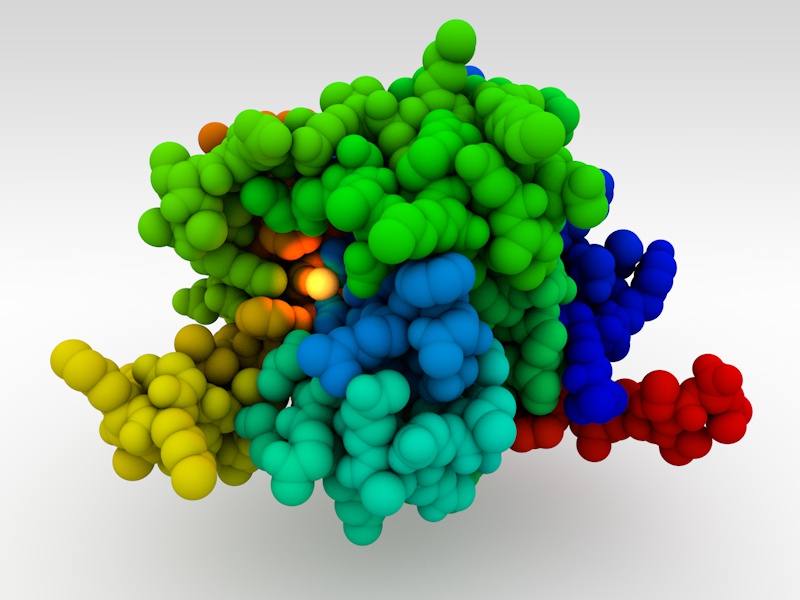
The World Health Organization just announced three recent deaths from Ebola in the Democratic Republic of Congo, which may grown into a new outbreak of the disease. Please support the Outsmart Ebola Together project and help scientists find better treatments for this deadly virus.
You can help researchers at The Scripps Research Institute find a cure for Ebola by donating your computing power to this project and encouraging others to join.
You can also support the research team by contributing to The Scripps Research Institute’s crowdfunding campaign. The team will use these funds to analyze the enormous volume of data generated by Outsmart Ebola Together and study the most promising drug candidates.
The Ebola virus is a significant global health threat and is a growing humanitarian crisis in Africa, killing thousands of victims in 2014.

http://www.webmd.com/a-to-z-guides/ebola-fever-virus-infection
If not handled properly, an Ebola outbreak can turn into an epidemic, overwhelming regional health services and disrupting trade and the delivery of social services, causing the welfare and economy of a region to deteriorate. The ongoing viral load in the human population increases the likelihood of further mutation. Additionally, the virus’s long incubation period and our highly connected modern world could allow the virus to spread to new geographies and across oceans.
Currently, there are no approved treatments or vaccines for this deadly disease, and the search for an effective antiviral drug to treat the disease is a high priority. While previous outbreaks have ended when the disease disappeared from the human population, the scope of the 2014 outbreak raises the possibility that the virus, rather than disappearing again, could become endemic – permanently persisting in human populations in one or more areas.
Outsmart Ebola Together on World Community Grid aims to help researchers at The Scripps Research Institute develop a treatment for Ebola virus. The computational power donated by World Community Grid volunteers is being used to screen millions of candidate drug molecules to identify ones that can disable the Ebola virus.
See the full article here.
Ways to access the blog:
https://sciencesprings.wordpress.com
http://facebook.com/sciencesprings
Please help promote STEM in your local schools.
![]()
“World Community Grid (WCG) brings people together from across the globe to create the largest non-profit computing grid benefiting humanity. It does this by pooling surplus computer processing power. We believe that innovation combined with visionary scientific research and large-scale volunteerism can help make the planet smarter. Our success depends on like-minded individuals – like you.”
WCG projects run on BOINC software from UC Berkeley.

BOINC is a leader in the field(s) of Distributed Computing, Grid Computing and Citizen Cyberscience.BOINC is more properly the Berkeley Open Infrastructure for Network Computing.
CAN ONE PERSON MAKE A DIFFERENCE? YOU BET!!
My BOINC

“Download and install secure, free software that captures your computer’s spare power when it is on, but idle. You will then be a World Community Grid volunteer. It’s that simple!” You can download the software at either WCG or BOINC.
Please visit the project pages-
Help Stop TB

Outsmart Ebola together
Discovering Dengue Drugs – Together


World Community Grid is a social initiative of IBM Corporation
IBM Corporation